DNA Methylation Analysis
DNA Methylation Analysis
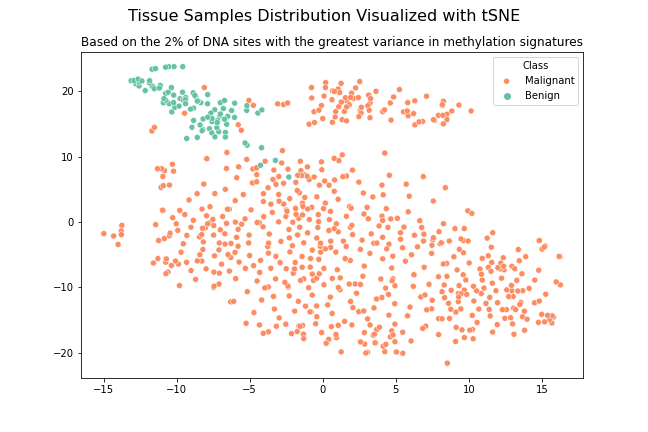
This project aims to classify healthy tissue samples and cancer tumor samples using DNA methylation signature data sets. A comprehensive data cleaning and processing methodology was implemented to fill missing values and create a subset of high variance DNA sites. A range of supervised machine learning models, including Logistic Regression, Decision Tree, Random Forest, and Extra Tree, were applied and evaluated. The Random Forest model achieved the highest accuracy. Unsupervised clustering models such as KNN and t-Distributed Stochastic Neighbor Embedding (t-SNE) were also utilized as a supplementary method to verify the results. The t-SNE model not only returned the desired class clusters but also provided additional insights of interest. As a next step, the feature importance of the Random Forest model will be further examined and the t-SNE clustering model will be explored in more detail. These findings have the potential to contribute to the development of new cancer tests and treatments utilizing DNA methylation signatures.